**Screening Antibacterial Activity: A Crucial Step in Drug Discovery**
Commentry - (2024) Volume 13, Issue 3
Description
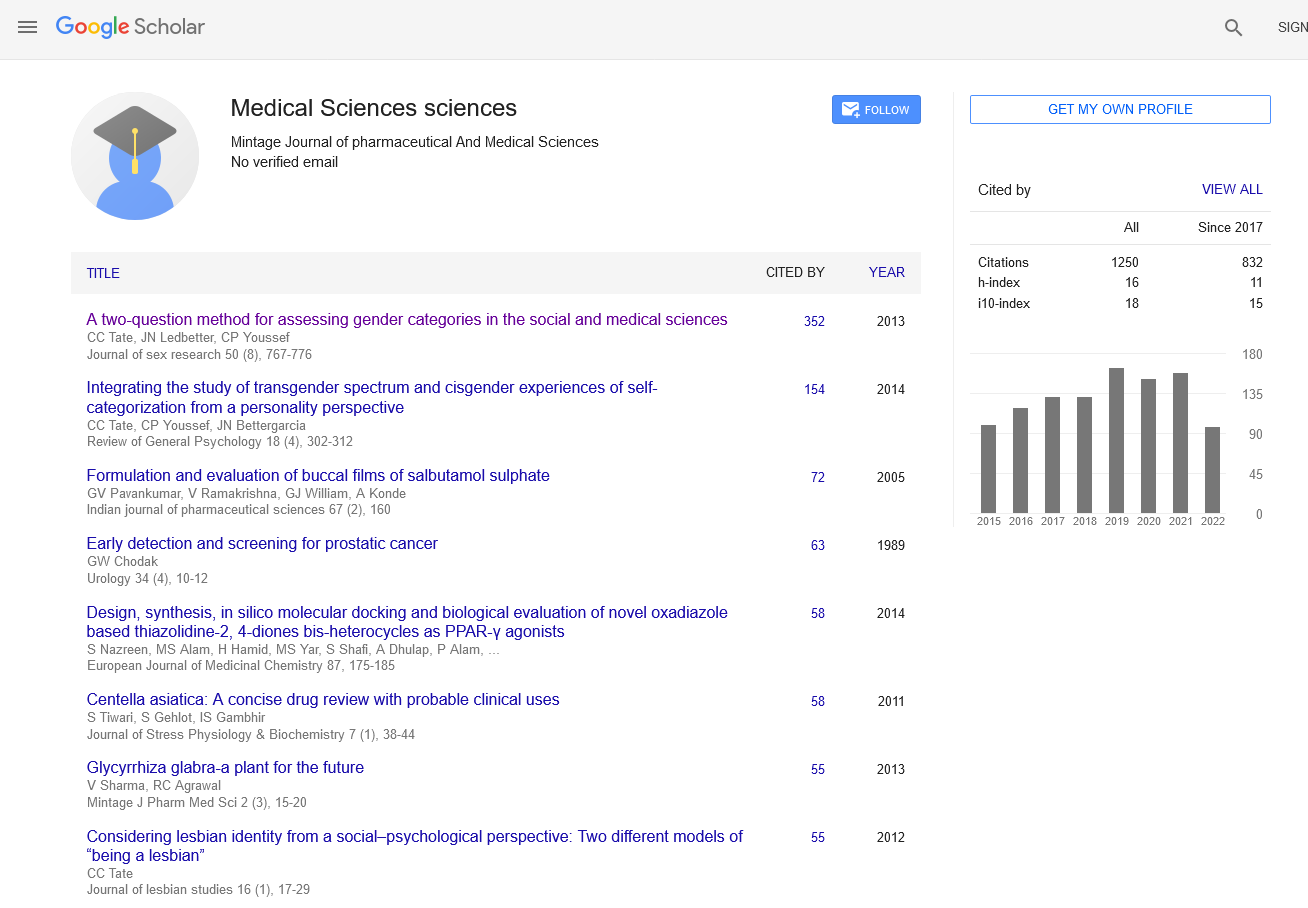
Screening for antibacterial activity is a fundamental process in the development of new antibiotics and the evaluation of substances that can inhibit bacterial growth or kill harmful bacteria. This process plays a crucial role in addressing the ongoing global challenge of antibiotic resistance, as researchers continuously search for new antibacterial agents capable of combatting resistant strains. Screening is a multi-step approach that involves identifying potential antibacterial compounds, testing them against specific bacterial strains, and evaluating their efficacy, toxicity, and potential as drug candidates. The first step in screening for antibacterial activity involves the selection of compounds to be tested. These compounds can come from a variety of sources, including natural products, synthetic chemicals, and libraries of previously untested substances. Natural products, such as plant extracts, microbial metabolites, and marine organisms, have historically been a rich source of antibiotics. For example, penicillin, discovered from the mold Penicillium notatum, remains one of the most successful antibiotics in history. Many researchers continue to explore natural environments in search of new bioactive compounds with antibacterial properties. Once a pool of potential compounds has been selected, the next step is to test them against bacterial strains of interest. The most commonly used method for initial screening is the agar diffusion method, which includes techniques such as the disk diffusion assay or the well diffusion assay. In these methods, bacterial cultures are spread onto an agar plate, and small disks or wells containing the test compounds are placed on the surface. The compounds diffuse into the agar, and after a period of incubation, the bacterial growth around the disks or wells is observed. If a compound has antibacterial activity, it will create a clear zone of inhibition around the disk or well, indicating that the bacteria in that area have been killed or prevented from growing. Another widely used technique for screening antibacterial activity is the broth dilution method, which can be performed in liquid media. In this method, bacteria are grown in a series of test tubes or wells containing different concentrations of the compound being tested. The bacteria are incubated with the compound, and their growth is monitored over time. This method helps determine the Minimum Inhibitory Concentration (MIC), which is the lowest concentration of the compound that prevents visible bacterial growth. The MIC is a key parameter in evaluating the potency of an antibacterial agent and is often used to compare the efficacy of different compounds. High-throughput screening (HTS) is another approach commonly used to screen large libraries of compounds for antibacterial activity. This automated process allows researchers to rapidly test thousands or even millions of compounds against bacterial strains in a relatively short amount of time. HTS relies on advanced robotics, microplate readers, and computational software to evaluate the antibacterial effects of the tested compounds. The ability to screen such vast numbers of compounds quickly makes HTS an invaluable tool in drug discovery, enabling the identification of promising lead compounds that can be further developed. In addition to evaluating the antibacterial efficacy of a compound, screening methods also assess the compound’s selectivity and toxicity. It is important that an antibacterial agent targets harmful bacteria without affecting human cells or beneficial microbes. Tests are conducted to ensure that the compounds are non-toxic to mammalian cells and do not disrupt the natural microbiota, as broad-spectrum antibiotics can sometimes cause unintended harm to beneficial bacteria in the body. Selectivity assays and cytotoxicity tests help researchers identify compounds with high therapeutic potential and minimal side effects. Following initial screening, the most promising compounds undergo further testing to determine their mechanism of action. Understanding how a compound exerts its antibacterial effects is essential for drug development, as it provides insight into how the compound interacts with bacterial cells. Some compounds work by targeting the bacterial cell wall, such as beta-lactam antibiotics, while others inhibit protein synthesis, DNA replication, or other critical cellular functions. Techniques like flow cytometry, electron microscopy, and genomic sequencing can help identify these mechanisms, guiding the optimization of lead compounds and improving their efficacy. In conclusion, screening for antibacterial activity is a critical step in the discovery and development of new antibiotics. With the rise of antibiotic resistance posing a serious threat to global public health, the need for novel antibacterial agents has never been more urgent. Through a combination of traditional techniques, high-throughput screening, and advanced mechanistic studies, researchers continue to identify promising compounds with the potential to combat bacterial infections. The ongoing search for new antibiotics, coupled with innovative screening methods, offers hope in addressing the growing challenge of bacterial resistance and ensuring the availability of effective treatments for future generations.
Acknowledgement
The authors are very thankful and honoured to publish this article in the respective Journal and are also very great full to the reviewers for their positive response to this article publication.
Conflict Of Interest
We have no conflict of interests to disclose and the manuscript has been read and approved by all named authors.
Author Info
Fernandez Vactor*Received: 02-Sep-2024, Manuscript No. mjpms-24-147874; , Pre QC No. mjpms-24-147874 (PQ); Editor assigned: 04-Sep-2024, Pre QC No. mjpms-24-147874 (PQ); Reviewed: 18-Sep-2024, QC No. mjpms-24-147874; Revised: 23-Sep-2024, Manuscript No. mjpms-24-147874 (R); Published: 30-Sep-2024, DOI: 10.4303/2320-3315/236026
Copyright: © This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/)

ISSN: 2320-3315
ICV :81.58